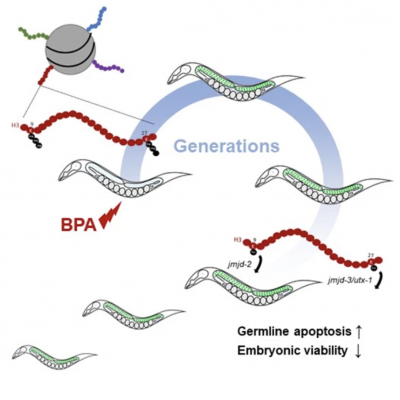
This project explores the environmental influences on the germline epigenome and their effects over multiple generations. Led by ISG faculty Patrick Allard, researchers at the UCLA Allard Lab are taking advantage of various innovative models and genetic tools to detect chemical disruption of the germline chromatin and in particular of evolutionary conserved epigenetic marks such as histone modifications. Through the collection of assays developed in the Allard Lab, they can investigate the multi- and trans-generational transmission of altered chromatin states originating from a single generational exposure. With this research, the Allard Lab not only identify the chemicals that affect the epigenome but also characterize their mechanism of action on epigenetic pathways. Some of their key publications on the topic are linked below:
Camacho J, Truong L, Kurt Z, Chen YW, Morselli M, Gutierrez G, Pellegrini M, Yang X, Allard P. The Memory of Environmental Chemical Exposure in C. elegans Is Dependent on the Jumonji Demethylases jmjd-2 and jmjd-3/utx-1. Cell Reports, Volume 23, Issue 8, 2392 – 2404
Camacho J, Allard P. Histone Modifications: Epigenetic Mediators of Environmental Exposure Memory. Epigenet Insights. 2018 Oct 14;11:2516865718803641. doi: 10.1177/2516865718803641. eCollection 2018. PMID: 30443644
Lundby Z, Camacho J, Allard P. Fast functional germline and epigenetic assays in the nematode Caenorhabditis elegans. In High Throughput Toxicity Assays: Methods and Protocols. Springer 2016. Chapter.


